EPPES parameter estimation example using Lorenz 95
This example demonstrates EPPES method using so called stochastic Lorenz 95 system [Wilks, 2005]. It is a Lorenz 95 system with added fast state variables whose effect is parameterized in the simpler forecast model. Here we estimate these parameters using EPPES. See the EPPES article [Laine, 2011] for details. This code is a slightly simplified version of the example in Section 4 of the paper.
Laine, et al.: Ensemble prediction and parameter estimation system: the method, Q. J. R. Meteorol. Soc., 2011, doi:10.1002/qj.922.
First, we load the truth system used as observations. The true states are generated with the full system using short integration time step and including the fast state variables, also. For estimation of the closure parameters of the simpler forecast model some independent random noise (with std sigma_a) is later added to these error free states.
T10 = load('T10.mat') truth = T10.truth; % true states aint = T10.aint; % analysis intervall dt = T10.dt; % integration time step
T10 =
truth: [301x360 double]
aint: 0.4
dt: 0.0025
The forecast model function is obtained by evaluating the numerical solution of the L95 ode system at given time points and initial values at the start. The modified Lorenz 95 ode system is defined in file the l95ode.m.
l95fun = @(time,s0,params) deval(ode45(@l95ode,time,s0(:),[],params),time);
The standard deviations for analysis and initial perturbations.
sigma_a = 0.05; sigma_ip = 0.1;
Forecast model settings.
params.F = 10; params.sigma_e = 0; params.phi = 0.0; params.gupars = [0.0 0.2]; % initial parameter values params.gufun = @(x,p) polyval(p,x); params.eta = zeros(50,1); fc_range = 3; % how long ahead do we forecast nens = 50; % ensemble size npar = length(params.gupars); % number of parameters
Initial values for hyperprior parameters.
mu = params.gupars(:); % initial prior mean tau = 100*diag([1;1].^2); % accuracy of mu sig = 0.1*diag([1;1].^2); % initial prior covariance n0 = 1; % accuracy of sig nmax = 10; % maximum n0
Define number of EPPES iterations, and allocate memory for saving the intermediate results,
niter = size(truth,1)-fc_range-1; niter = 30; % use a smaller number of iterations for faster illustration parsall = zeros(niter,nens,npar); weightsall = zeros(niter,nens); muall = zeros(niter,npar); sigall = zeros(niter,npar,npar); disp('running eppes, this will take some time ...')
running eppes, this will take some time ...
The simulation loop.
figure(1);clf for iiter=1:niter time = (iiter-1)*aint:dt:(iiter+fc_range-1)*aint; a = truth(iiter,1:40); a = a + sigma_a*randn(size(a)); % analysis = truth + noise pars_prop = mvnorr(nens,mu,sig); % proposed parameters from N(mu,sig) % launch the ensemble for j=1:nens s0 = a + sigma_ip*randn(size(a)); % initial value params.gupars = pars_prop(j,:); % forecast model parameters sens(:,:,j) = l95fun(time,s0,params)'; % run the forecast model smod = sens(aint/dt:aint/dt:end,:,:); % model response at data points end % sum of squares for ensemble members is calculated in function ens_ss_l95.m ss = ens_ss_l95(smod,truth(iiter+1:iiter+fc_range,1:40)); % calculating weights and resampling w = exp(-0.5*(ss(:))); w = w/sum(w); pars_new = resample(pars_prop,w); % update the parameters [mu,tau,nn,sig] = eppesupd(pars_new,mu,tau,n0,sig); n0 = min(nn,nmax); % parameter n0 is at most nmax % save the proposed points and their resample weights parsall(iiter,:,:) = pars_prop; weightsall(iiter,:) = w'; muall(iiter,:) = mu'; sigall(iiter,:,:) = sig; % some plotting on the fly figure(1); subplot(3,1,1); colormap(flipud(gray)); plotc(repmat(time,nens,1),squeeze(sens(:,1,:))',-ss); hold on; plot(time(1)+aint:aint:time(end),truth(iiter+1:iiter+fc_range,1),'ro',... 'markersize',10,'markerfacecolor','red'); hold off; xlabel('time'); ylabel('state X_1'); title(sprintf('iter: %d/%d',iiter,niter)); subplot(3,1,2); plotc(pars_prop(:,1),pars_prop(:,2),w,'o','markeredgecolor',[0.85 0.85 0.85]); hold on ellipse(mu,4.6*sig,'g-','LineWidth',2); plot(mu(1),mu(2),'.k'); hold off xlabel('parameter \theta_1'); ylabel('parameter \theta_2'); subplot(3,1,3); stem(w); xlabel('ensemble members'); ylabel('parameter weight') drawnow; end
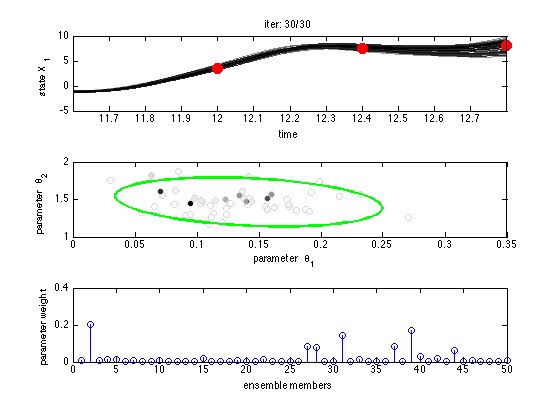
The figure shows iteratively the state evolution of each ensemble member for the first state variable X1, together with the observations. The forecast model lines are colored according to the forecast skill weight (upper panel). Two dimensional plot of the proposed parameters with their weights are shown with 95% probability ellipsoid of the proposal/prior covariance sig (center panel). The calculated relative weight for each members are shown (lower panel).
disp('... done')
... done
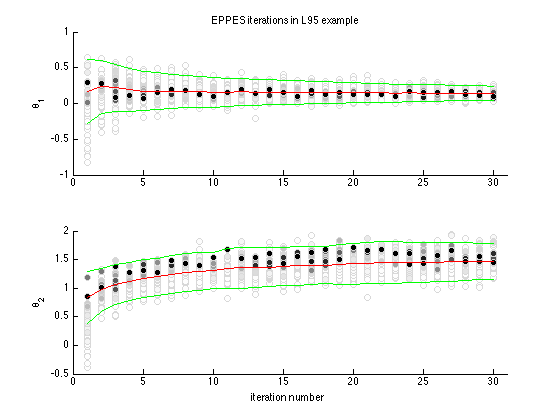
Plot the proposed points and their weights. For both of the parameters theta1 and theta2 the evolution of the EPPES method is shown. Iteration number is on the x axis and parameter value at y axis. Colouring shows the importance weights, red line is the evolution of the mean mu and green lines show plus minus 2 times the standard deviation from covariance matrix sig.
figure(2); clf subplot(2,1,1) eppeschainplot(parsall(:,:,1),weightsall, muall(:,1), sqrt(sigall(:,1,1))) ylabel('\theta_1') title('EPPES iterations in L95 example') subplot(2,1,2) eppeschainplot(parsall(:,:,2),weightsall, muall(:,2), sqrt(sigall(:,2,2))) ylabel('\theta_2') xlabel('iteration number')