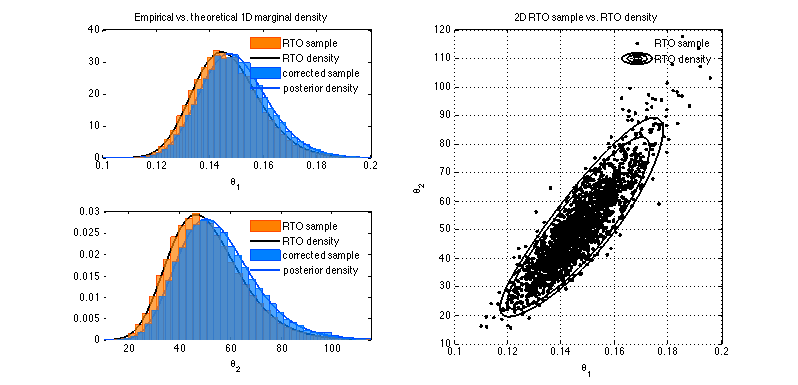
2D RTO example Monod model
We sample form the posterior of two parameters of the Monod model
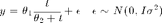
given the data.
Data and initial values
x = [28, 55, 83, 110, 138, 225, 375]'; y = [0.053, 0.060, 0.112, 0.105, 0.099, 0.122, 0.125]'; th = [0.15,50]; % initial theta sig = 0.01; % known observation error std
Model function, Jacobian, sum of squares
f = @(x,th) th(1).*x./(th(2) + x); j = @(x,th) [x(:)./(th(2)+x(:)),-th(1).*x(:)./(th(2)+x(:)).^2]; ss = @(th,data) sum(((y-f(x,th))./sig).^2);
Initial fit and Q from qr decomposition of Jacobian at MAP
th = lsqlevmar(@(th)(y-f(x,th))/sig,@(th)j(x,th)/sig,th); [Q0,~] = qr(j(x,th)/sig,0);
RTO sampling using Levenberg - Marquardt
nsimu = 20000; thsim = zeros(nsimu,2); % rto sample rtoc = zeros(nsimu,1); % log correction factor h = waitbar(0,'Doing RTO simulation...'); for i=1:nsimu if fix(i/500) == i/500;waitbar(i/nsimu,h);end ys = y+randn(size(y))*sig; % randomize % then optimize thsim(i,:) = lsqlevmar(@(th)Q0'*(ys-f(x,th))/sig,@(th)Q0'*j(x,th)/sig,th); % log of the correction factor rtoc(i) = rtodetlog(j(x,thsim(i,:))/sig,(y-f(x,thsim(i,:)))./sig,Q0); end delete(h);
Resample to correct the sample
%thsimfix = logresample(thsim,2*rtoc); % Importance sampling thsimfix = thsim(rtoaccept_log(rtoc),:); % M-H sampling
Calculate the true posterior distribution over a mesh
ng = 100; CC = zeros(ng); % correction factors PP = zeros(ng); % true posterior th1 = linspace(0.10,0.2,ng); th2 = linspace(10,115,ng); [TH1,TH2] = meshgrid(th1,th2); cin = @(x,l1,u1, l2,u2) sum( x(:,1)>l1 & x(:,1)<=u1 & x(:,2)>l2 & x(:,2)<=u2 ); d1 = th1(2)-th1(1); d2 = th2(2)-th2(1); for ji=1:ng for ii=1:ng thi = [TH1(ii,ji),TH2(ii,ji)]; CC(ii,ji) = exp(rtodetlog(j(x,thi)./sig,(y-f(x,thi))./sig,Q0)); PP(ii,ji) = exp(-0.5*ss(thi,[])); end end RR = CC.*PP; RR = RR./sum(sum(RR)); % RTO distribution PP = PP./sum(sum(PP)); % posterior distribution
Plotting
figure(1); clf % theta_1 marginal subplot(2,2,1) nhist = 50; h = histp(thsim(:,1),nhist); set(h,'facecolor',[1 0.5 0],'edgecolor',[1 0.3 0]); set(get(h,'Children'),'Facealpha',0.7); hold on h = plot(th1,sum(RR)./sum(sum(RR))/d1,'-','color',[0 0 0],'linewidth',2); % posterior h = histp(thsimfix(:,1),nhist); set(h,'facecolor',[0 0.5 1],'edgecolor',[0 0.3 1]); set(get(h,'Children'),'Facealpha',0.7); plot(th1,sum(PP)./sum(sum(PP))/d1,'-','color',[0 0.3 1],'linewidth',2); hl = legend('RTO sample','RTO density','corrected sample','posterior density'); set(hl,'box','off') hold off xlabel('\theta_1') title('Empirical vs. theoretical 1D marginal density'); xlim(th1([1 end])); % theta_2 marginal subplot(2,2,3) h = histp(thsim(:,2),nhist); set(h,'facecolor',[1 0.5 0],'edgecolor',[1 0.3 0]); set(get(h,'Children'),'Facealpha',0.7); hold on h = plot(th2,sum(RR')./sum(sum(RR'))/d2,'-','color',[0 0 0],'linewidth',2); % posterior h = histp(thsimfix(:,2),nhist); set(h,'facecolor',[0 0.5 1],'edgecolor',[0 0.3 1]); set(get(h,'Children'),'Facealpha',0.7); plot(th2,sum(PP')./sum(sum(PP'))/d2,'-','color',[0 0.3 1],'linewidth',2); hold off hl = legend('RTO sample','RTO density','corrected sample','posterior density'); set(hl,'box','off') xlabel('\theta_2') xlim(th2([1 end])); % 2D density subplot(2,2,[2 4]) plot(thsim(1:10:end,1),thsim(1:10:end,2),'.k') % contour limits p1 = plims2dgrid(RR,[0.5 0.9 0.95]); hold on contour(th1,th2,RR,p1,'-k','linewidth',2); hold off xlim([0.1 0.2]) ylim([10 120]) grid xlabel('\theta_1') ylabel('\theta_2') title('2D RTO sample vs. RTO density') hl = legend('RTO sample','RTO density'); set(hl,'box','off')